
Never-before-seen forms and unexpected connections among proteins revealed by analysis of their shapes.
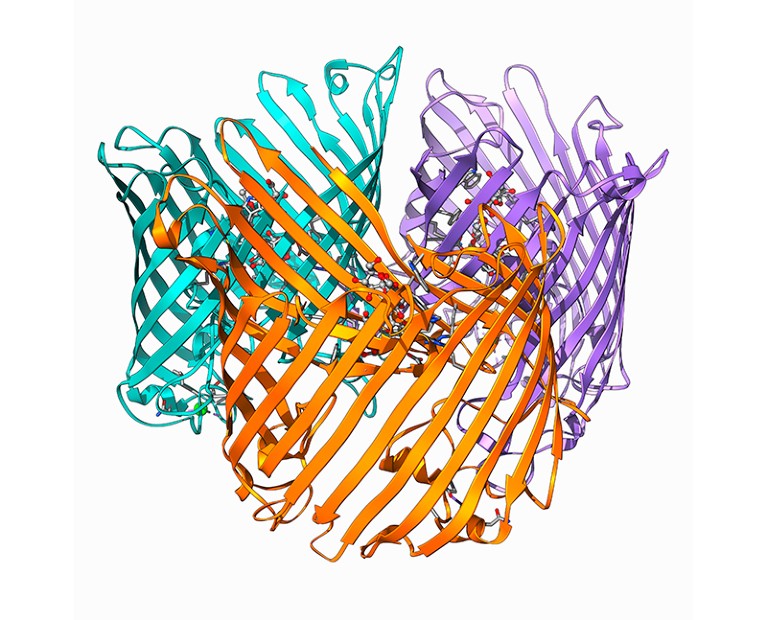
A protein shape called a Beta-barrel.Credit: Laguna Design/Science Photo Library
The protein universe just got a lot brighter.
Researchers have mined a database containing the structures of nearly every known protein — more than 200 million entries predicted using Google DeepMind’s revolutionary AlphaFold neural network. The work has uncovered completely new shapes, surprising connections in the machinery of life, and other insights that would have been unthinkable a few years ago.
Foldseek gives AlphaFold protein database a rapid search tool
“Thanks to AlphaFold we can now explore entire families of proteins we knew nothing about,” says Eduard Porta Pardo, a computational biologist at Josep Carreras Leukaemia Research Institute (IJC) in Barcelona, Spain, who was not involved in a pair of studies published1,2 on 13 September in Nature.
Last year, Google DeepMind used AlphaFold to predict the structure of nearly every known protein from organisms with genome data, amassing some 214 million structures in the AlphaFold database, which is hosted by the European Molecular Biology Laboratory’s European Bioinformatics Institute (EMBL-EBI) in Hinxton, UK.
Scientists found the resource instantly handy, but many of them looked only at a single structure, or family of related structures, says Martin Steinegger, a computational biologist at Seoul National University, who was interested in mapping the relationships of the entire database. “I thought it would be interesting to see how big our structural universe really is.”
To do this, a team co-led by Steinegger and computational biologist Pedro Beltrao, at ETH Zurich in Switzerland, developed a tool that quickly could compare every structure in the database, based on similarities in their shape. This identified more than 2 million ‘clusters’ of similarly shaped proteins in the AlphaFold database1.
Researchers have conventionally made such comparisons using protein sequences, which are encoded by genes. But protein sequences tend to change more rapidly over evolutionary time, compared with their structures, limiting the ability to find very distantly related proteins. Steinegger estimates that by comparing protein structures, they have identified 10 times the number of clusters of related proteins than they would using only sequences.
The researchers have only begun to explore these newly identified ‘galaxies’ in the protein universe, but they have already turned up some surprising connections. For instance, they found that a protein that humans and other complex organisms use to detect viral DNA and trigger a quick immune attack is in a cluster with proteins from single-celled bacteria and archaea — a link that wasn’t known before, says Steinegger.
Next to nothing is known about more than one-third of the protein clusters. “I really hope that biologists put some light on this darkness,” Steinegger says.
A second team took a slightly different approach to illuminating the dark matter of the protein universe. Computational biologists Joana Pereira, Janani Durairaj, Torsten Schwede, at the University of Basel in Switzerland and the SIB Swiss Institute of Bioinformatics, and their colleagues created a network that connected more than 50 million of the most accurately predicted structures in the AlphaFold database (the tool provides a measure of how good it thinks its predictions are). They then used these groupings to identify some of the darkest corners of the protein universe2.
One pleasant surprise was a protein shape that had never been seen before. The researchers dubbed it a ‘Beta-flower’ because the structures contain a number of hairpin turns — found in a known protein shape called a Beta-barrel — that resemble petals on a flower. Proteins that contain Beta-flowers are distantly related to one another, but it’s not clear what they do, says Pereira, who is studying the shape further.
“This work actually opened up a Pandora’s box of projects. We have to decide which to prioritize,” Pereira adds. She and her colleagues hope other researchers use their network to see how their favourite protein fits into the wider universe of molecules.
Christine Orengo, a computational biologist at University College London, is excited to have new ways of navigating the protein universe. But she cautions that some AlphaFold predictions deemed to be highly accurate for an entire protein may less accurately represent the shapes of functional portions of the protein, or domains, that researchers are interested in. Leaving those anomalies aside should still leave researchers with a treasure trove of new protein families, Orengo says, “which is incredibly exciting.”
